Difference between revisions of "Google Summer of Code Ideas"
Sabareeshs (talk | contribs) |
|||
| Line 18: | Line 18: | ||
MSL has been extensively used in scientific publications encompassing a variety of applications. For example, [http://senes.biochem.wisc.edu/pdf/FtsBdimer-preprint.pdf protein structure prediction], [http://www.ncbi.nlm.nih.gov/pubmed/23422424 protein design], [http://www.ncbi.nlm.nih.gov/pubmed/23089864 protein engineering] and [http://senes.biochem.wisc.edu/pdf/EBL-2012-Proteins.pdf modeling algorithms/methods], and others. | MSL has been extensively used in scientific publications encompassing a variety of applications. For example, [http://senes.biochem.wisc.edu/pdf/FtsBdimer-preprint.pdf protein structure prediction], [http://www.ncbi.nlm.nih.gov/pubmed/23422424 protein design], [http://www.ncbi.nlm.nih.gov/pubmed/23089864 protein engineering] and [http://senes.biochem.wisc.edu/pdf/EBL-2012-Proteins.pdf modeling algorithms/methods], and others. | ||
| + | ==Current development== | ||
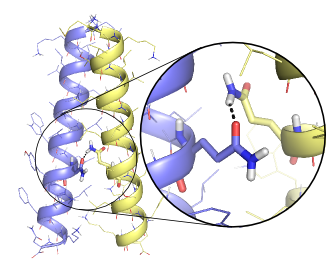
| − | + | [[File:FtsB.png | frame | '''Structural model of a dimer of a membrane protein obtained with MSL''']] | |
| + | |||
| + | The main theme of MSL development will continue to be the creation of a flexible yet powerful software framework that is easily accessible to programmers familiar with C++/object-orientation. | ||
| − | + | We are striving to improve the core framework through | |
* more advanced objects | * more advanced objects | ||
* more efficient implementations | * more efficient implementations | ||
| Line 34: | Line 37: | ||
There are a number of areas, listed below, in which a GSoC student could contribute to realize these goals. | There are a number of areas, listed below, in which a GSoC student could contribute to realize these goals. | ||
| + | |||
| + | |||
== Ideas == | == Ideas == | ||
Revision as of 18:43, 1 April 2013
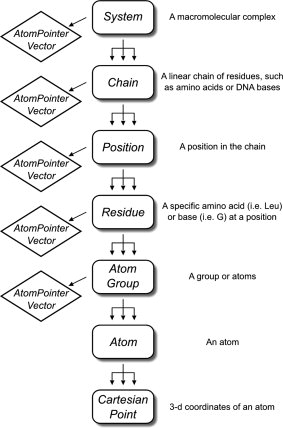
MSL (Molecular Software Libraries) is an open source C++ library of objects related to macromolecular modeling.
Contents
Goal
The goal of MSL is to create a toolbox to enable a programmer to design powerful and advanced applications for molecular analysis, modeling, prediction and design. At the same time, MSL should also allow the rapid creation of applications that perform simple tasks (for example, measuring the distance between two atoms).
History
MSL has been developed in the past 5 years (9 years if we include its early predecessor). It has an extensive code base (over 150 objects and 100,000 lines of code). After years in beta, the first stable version (1.0) was released in 2012.
The code can be downloaded at the MSL SourceForge page and the entire development tree is available at the MSL SVN repository. Several examples are now available in the repository that demonstrate the capabilities of MSL objects in modeling applications.
MSL is described in its primary citation:
- Kulp DW, Subramaniam S, Donald JE, Hannigan BT, Mueller BK, Grigoryan G and Senes A "Structural informatics, modeling and design with an open-source Molecular Software Library (MSL)", J Comput. Chem. 2012 33(20), 1645-61 (Download PDF)
MSL has been extensively used in scientific publications encompassing a variety of applications. For example, protein structure prediction, protein design, protein engineering and modeling algorithms/methods, and others.
Current development
The main theme of MSL development will continue to be the creation of a flexible yet powerful software framework that is easily accessible to programmers familiar with C++/object-orientation.
We are striving to improve the core framework through
- more advanced objects
- more efficient implementations
- implementing more modeling algorithms and protocols
Another important direction is making MSL more accessible to a wider audience by
- creating interfaces to other programming languages
- making distribution of MSL easier
- building ready-to-use applications
- hosting MSL applications on web servers for public use
There are a number of areas, listed below, in which a GSoC student could contribute to realize these goals.
Ideas
Embedding MSL code for use in higher-level languages
Brief explanation:
The idea is to make MSL code available to programs in other higher-level languages. This could be done using an interface compiler like the SWIG (see swig.org)
Expected results:
Infrastructure and example applications in multiple higher-level languages.
Knowledge Prerequisite:
C++ and one of Python or R or perl
MSL-Light
Brief explanation:
The objects in MSL are sometimes too complex for simple operations on large molecules. The idea is to implement light-weight versions of some core objects, for example, the AtomContainer object is a lighter version of the System object.
Expected results:
A collection of light-weight objects and methods.
Knowledge Prerequisite:
C++, Data Structures and Algorithms
Smart pointers in MSL
Brief explanation:
MSL uses basic pointers which are prone to bugs due to dangling pointers, memory leaks, etc.,
Expected results:
Replace regular pointers with the appropriate smart pointers in MSL.
Knowledge Prerequisite:
C++
Efficient program options management
Brief explanation:
MSL has an advanced OptionParser object that supports name/value based program options. A mechanism needs to be implemented to group program options for MSL that uses the underlying OptionParser.
Expected results:
The mechanism developed should facilitate option reuse and in general, easy option handling in MSL applications.
Knowledge Prerequisite:
C++
MSL Linux Distribution Packages
Brief explanation:
The MSL needs to be compiled from source. The distribution of the software as a .deb and/or .rpm package will simplify installation in some of the most popular Linux distros.
Expected results:
One-click/command MSL installation.
Knowledge Prerequisite:
Linux, Make
Developing a website to run MSL applications
Brief explanation:
There are several MSL applications that are directly useful to structural biologists. For example, the side-chain modeling programs, the structure prediction programs, etc., are of general interest and it would be useful to create a web interface that allows the use of these applications with a minimal/no knowledge of MSL.
Expected results:
A website to run MSL applications on the lab cluster, post process the output and make the results available to the user.
Knowledge Prerequisite:
Scripting Languages and Web Design
Contact
Please email sabareeshs@gmail.com if you are interested in working on any of these ideas and would like to obtain more information.